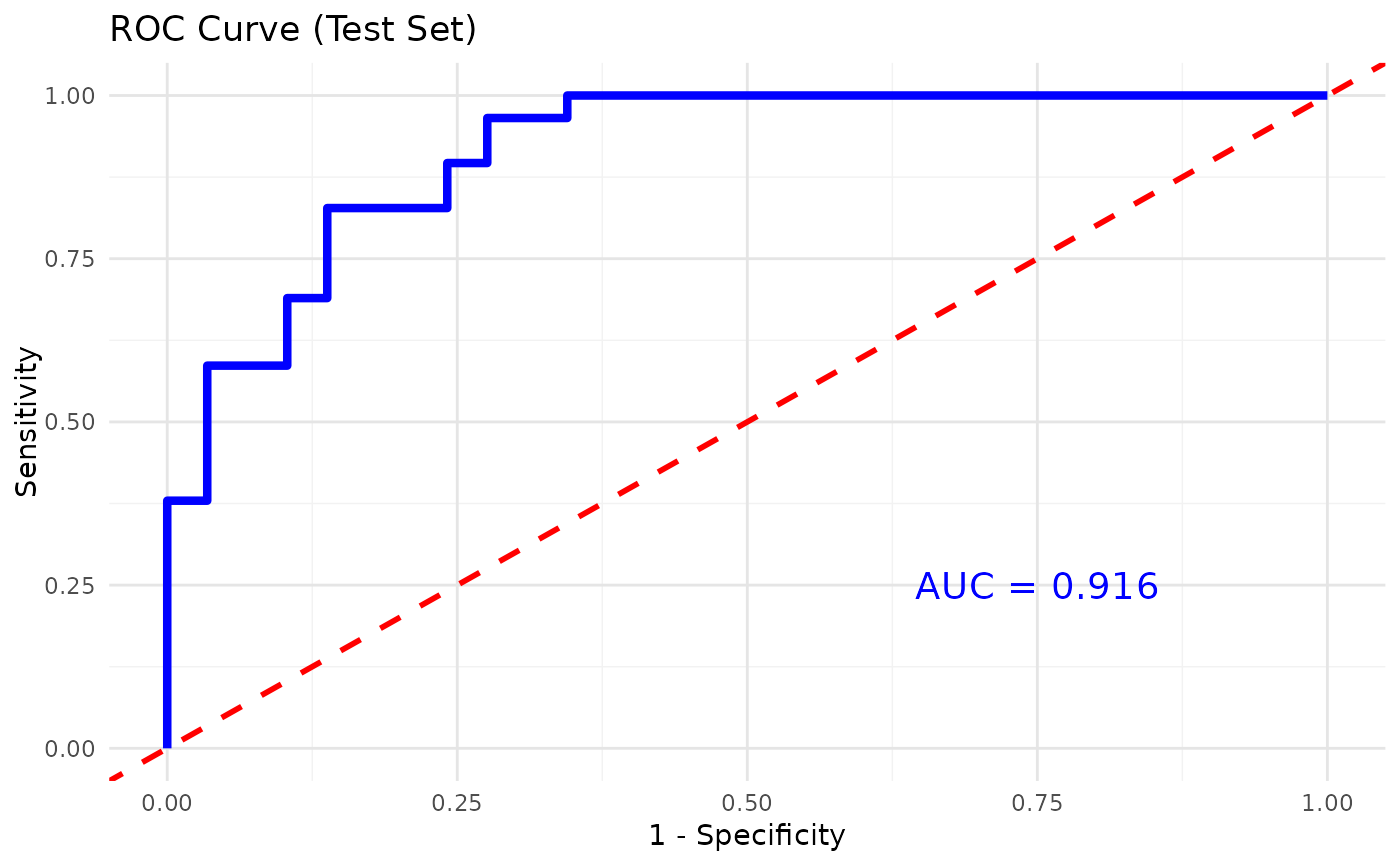
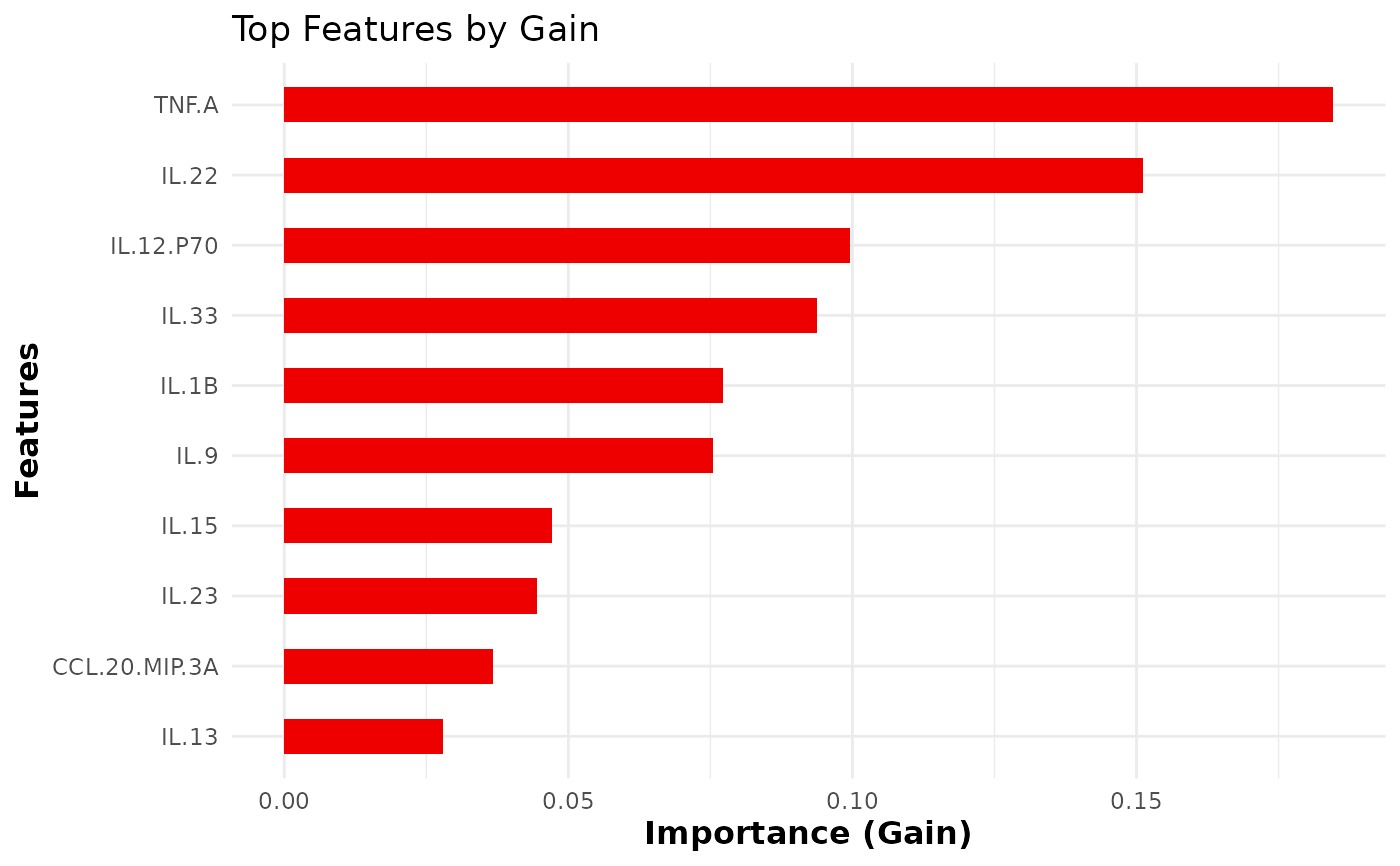
This function trains and evaluates an XGBoost classification model on cytokine data. It allows for hyperparameter tuning, cross-validation, and visualizes feature importance.
Usage
cyt_xgb(
data,
group_col,
train_fraction = 0.7,
nrounds = 500,
max_depth = 6,
eta = lifecycle::deprecated(),
learning_rate = 0.1,
nfold = 5,
cv = FALSE,
objective = "multi:softprob",
early_stopping_rounds = NULL,
eval_metric = "mlogloss",
gamma = lifecycle::deprecated(),
min_split_loss = 0,
colsample_bytree = 1,
subsample = 1,
min_child_weight = 1,
top_n_features = 10,
verbose = 1,
plot_roc = FALSE,
print_results = FALSE,
seed = 123
)Arguments
- data
A data frame containing the cytokine data, with one column as the grouping variable and the rest as numerical features.
- group_col
A string representing the name of the column with the grouping variable (i.e., the target variable for classification).
- train_fraction
A numeric value between 0 and 1 representing the proportion of data to use for training (default is 0.7).
- nrounds
An integer specifying the number of boosting rounds (default is 500).
- max_depth
An integer specifying the maximum depth of the trees (default is 6).
- eta
- learning_rate
A numeric value representing the learning rate (default is 0.1). This replaces the deprecated
etaargument.- nfold
An integer specifying the number of folds for cross-validation (default is 5).
- cv
A logical value indicating whether to perform cross-validation (default is FALSE).
- objective
A string specifying the XGBoost objective function (default is "multi:softprob" for multi-class classification).
- early_stopping_rounds
An integer specifying the number of rounds with no improvement to stop training early (default is NULL).
- eval_metric
A string specifying the evaluation metric (default is "mlogloss").
- gamma
- min_split_loss
A numeric value for the minimum loss reduction required to make a further partition (default is 0). This replaces the deprecated
gammaargument.- colsample_bytree
A numeric value specifying the subsample ratio of columns when constructing each tree (default is 1).
- subsample
A numeric value specifying the subsample ratio of the training instances (default is 1).
- min_child_weight
A numeric value specifying the minimum sum of instance weight needed in a child (default is 1).
- top_n_features
An integer specifying the number of top features to display in the importance plot (default is 10).
- verbose
An integer specifying the verbosity of the training process (default is 1).
- plot_roc
A logical value indicating whether to plot the ROC curve and calculate the AUC for binary classification (default is
FALSE).- print_results
A logical value indicating whether to print the results of the model training and evaluation (default is
FALSE). If set toTRUE, it will print the confusion matrix, and feature importance.- seed
An integer specifying the seed for reproducibility (default is 123).
Value
A list containing:
- model
The trained XGBoost model.
- confusion_matrix
The confusion matrix of the test set predictions.
- importance
The feature importance matrix for the top features.
- class_mapping
A named vector showing the mapping from class labels to numeric values used for training.
- cv_results
Cross-validation results, if cross-validation was performed (otherwise NULL).
- plot
A ggplot object showing the feature importance plot.
Details
The function allows for training an XGBoost model on cytokine data,
splitting the data into training and test sets. If cross-validation is
enabled (cv = TRUE), it performs k-fold cross-validation and reports the
confusion matrix and accuracy.
The function also visualizes the top N important
features using xgb.ggplot.importance().
Examples
# Example usage:
data_df0 <- ExampleData1
data_df <- data.frame(data_df0[, 1:3], log2(data_df0[, -c(1:3)]))
data_df <- data_df[, -c(2:3)]
data_df <- dplyr::filter(data_df, Group != "ND")
cyt_xgb(
data = data_df,
group_col = "Group",
nrounds = 500,
max_depth = 4,
min_split_loss = 0,
learning_rate = 0.05,
nfold = 5,
cv = FALSE,
objective = "multi:softprob",
eval_metric = "auc",
early_stopping_rounds = NULL,
top_n_features = 10,
verbose = 0,
plot_roc = TRUE,
print_results = FALSE)